Some of our ongoing projects
Hit one of the project buttons for more details
Project #1: DESIGN AND DEVELOPMENT OF FLOURESCENT PROTEIN BASED BIOSENSORS
Genetically encoded biosensors are indispensable tools for modern cell biology. Fluorescent protein-based live-cell imaging techniques have revolutionized our understanding of cellular processes. Almost any biological process can be investigated in detail with high spatial and temporal resolution using differently colored fluorescent protein variants. Nowadays, a plethora of genetic biosensors is available for more than 100 different targets including ions, metabolites, reactive oxygen, and reactive nitrogen species, temperature, pressure, signaling events, protein interactions and more. In the past, together with our collaboration partners, we have developed novel, or further refined existing Foerstner Resonance Energy Transfer (FRET)-based (Figure 1A), single FP based (Figure 1B) and differentially targeted biosensors (Figure 1C) for calcium, nitric oxide, ATP, potassium and pH levels. Prospective students are welcome to join us to develop novel and informative biosensors and imaging tools for (sub)cellular visualization of intracellular signaling events.
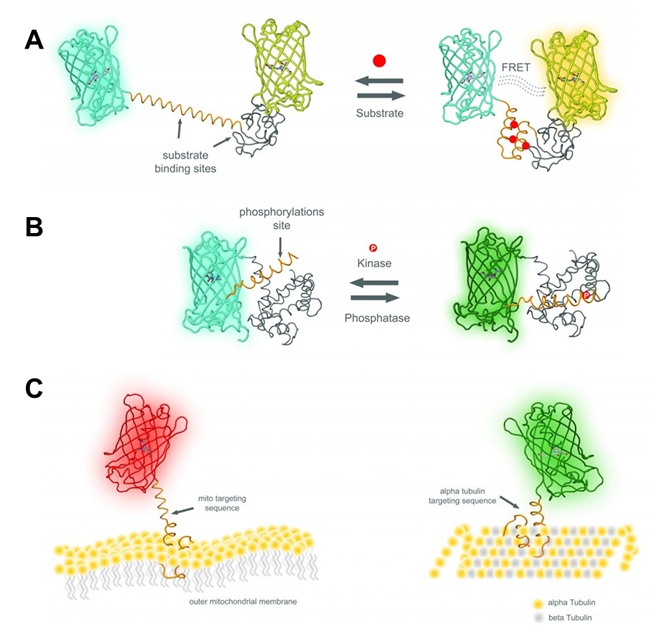
Figure 1: (A) Schematic of a classic FRET biosensor. A FRET-based sensor consists of two fluorescent proteins, a FRET donor (cyan FP) and a FRET acceptor (yellow FP). Both FPs are sandwich a substrate specific domain which is capable of binding the analyte of interest in a reversible manner (Calcium, ATP, etc.). In the presence of the analyte, the sensor domain of the construct undergoes a dramatic conformational rearrangement which permits the donor FP to transfer its energy to the acceptor which allows a ratiometric readout on the fluorescence microscope in real-time. (B) Schematic of a single FP based circularly permuted and ratiometric biosensor. This class of a sensor consists of only one fluorescent protein and two substrate specific domains which are capable of binding the analyte of interest in a reversible manner (Calcium, ATP, etc.). In the presence of the analyte, the sensor domains of the construct interact and lead to a dramatic conformational rearrangement of the fluorescent protein which in turn changes the spectral properties of the FP. This again allows a ratiometric readout on the fluorescence microscope in real-time. (C) Schematic of a differentially targeted FP to the outer mitochondrial membrane (left panel) and microtubule (right panel). Exploiting well-established and newly identified targeting peptides, we can direct genetic biosensors to specific subcellular locals within a cell or tissue. © Eroglu – 2016
Putative candidates joining this project will deal with:
Literature screen, identification, and assessment of a putative sensor or targeting element
in silico design and computation of biosensors using bioinformatics for 3-D modeling and calculation on the protein level.
Design and fabrication of DNA plasmids that includes standard molecular cloning approaches
Recombinant protein production, purification, and testing using fluorescent spectrophotometry
Cell culture methods for primary cells and various cancer cell lines
Purification of DNA-plasmids and transfection of cultured cells
Generation and application of recombinant viral vector systems including Adenoviral and Lentiviral systems
High-resolution fluorescence microscopy including Widefield and Confocal Microscopy
- Biostatistical Analysis, data interpretation, and presentation
Selected Literature for interested students:
Novel genetically encoded fluorescent probes enable real-time detection of potassium in vitro and in vivo. Bischof et al, 2017, Nature Communications
Project #2: MULTICHROMATIC IMAGING OF SIGNALING EVENTS IN INDIVIDUAL CELLS FROM THE VASCULATURE AND CANCER
Multiplex fluorescence imaging is a powerful approach with broad applications for in-vivo and single cell analysis. We exploit spectral variants of well-characterized biosensors. Thanks to their simple architecture and distinctive spectral properties, as well as their high selectivity and sensitivity for their particular analytes, both biosensors can be imaged simultaneously using standard fluorescence microscopy techniques. In this project, we will establish simultaneous imaging approaches with the geNOps biosensors, along with red shifted GECO indicators and red-shifted HyPer biosensors to investigate the relationship of Ca2+/H2O2, NO/Ca2+, and H2O2/NO pathways in response to various stimuli in healthy cells and disease states.

Figure 2: Schematic overview of chemogenetic approaches in vitro and in vivo and in cancer therapy.
Putative candidates joining this project will deal with:
Cell physiology of vascular cells and cancer cells with a particular focus on NO biosynthesis and ROS signaling
Design and development of bicistronic constructs for simultaneous expression of multiple biosensors
Multichromatic live cell imaging techniques using genetically encoded biosensors and chemical sensors for Ca2+, NO, and H2O2
Cell culture methods for primary cells and various cancer cell lines
Application of recombinant viral vector systems including Adenoviral and Lentiviral systems for the generation of stable cell lines
High-resolution confocal microscopy
- Biostatistical Analysis, data interpretation, and presentation
Selected Literature for interested students:
Discordance between eNOS phosphorylation and activation revealed by multispectral imaging and chemogenetic methods. Eroglu et al, 2019, PNAS
Project #3: A NOVEL CHEMOGENETIC APPROACH TO INVESTIGATE THE RELATIONSHIP OF REACTIVE OXYGEN SPECIES AND REACTIVE NITROGEN SPECIES IN SINGLE CELLS
The roles of ROS in biology continue to be discussed controversially despite decades of intensive investigation. For too long time ROS has been studied in the context of a damaging factor with a pathological impact on tissues and nucleic acids. The causative relationship between increases in intracellular ROS and the pathologies with which they are associated has remained unclear, owing to the lack of available tools to precisely and accurately manipulate ROS production both in vitro and in vivo. Yet a novel chemogenetic approach referred to as DAAO permits us to generate ROS levels in subcellular locals and tissues with excellent spatial resolution. DAAO stands for D-amino acid oxidase, which is yeast derived enzyme capable of converting D-amino acids to its corresponding L-amino acid generating equal concentrations of H2O2 during this process. Combination of DAAO constructs with relevant biosensors for specific analytes enable the generation and simultaneous detection of ROS mediated downstream effects. Moreover, differential targeting of this chimera into subcellular locales permits us to study the downstream effects of local ROS elevation on a variety of signaling events. We have a particular interest in exploring the roles of ROS on subcellular Ca2+ signaling, NO signaling and the energy metabolism of cells in response oxidative stress.

Figure 3: Multiparametric manipulation of local ROS levels and simultaneous imaging using different biosensors.
Putative candidates joining this project will deal with:
Cell physiology with a particular focus on ROS, RNS, and Energy metabolism in individual cells
Multiparametric imaging techniques with genetic and chemical biosensors along with chemogenetic tools
Agilent Seahorse XF Technology for real-time imaging of cellular energy metabolism
Single cell imaging of ATP levels using genetic biosensors (A-Team)
Application of recombinant viral vector systems including Adeno viral and Lentiviral systems
High-resolution confocal microscopy
small interfering molecules including siRNA and shRNA
- Biostatistical Analysis, data interpretation and presentation
Selected Literature for interested students:
Complexities of the chemogenetic toolkit: Differential mDAAO activation by d-amino substrates and subcellular targeting Erdoğan et al, 2021, FRBM
–
