Our ongoing projects
A Unifying Framework for Brain Microvascular Health
Life at the blood-brain barrier is a balancing act. Both reactive oxygen species (ROS) and intracellular pH are indispensable signalling cues, yet either parameter becomes detrimental when it drifts outside a narrow “Goldilocks” zone.
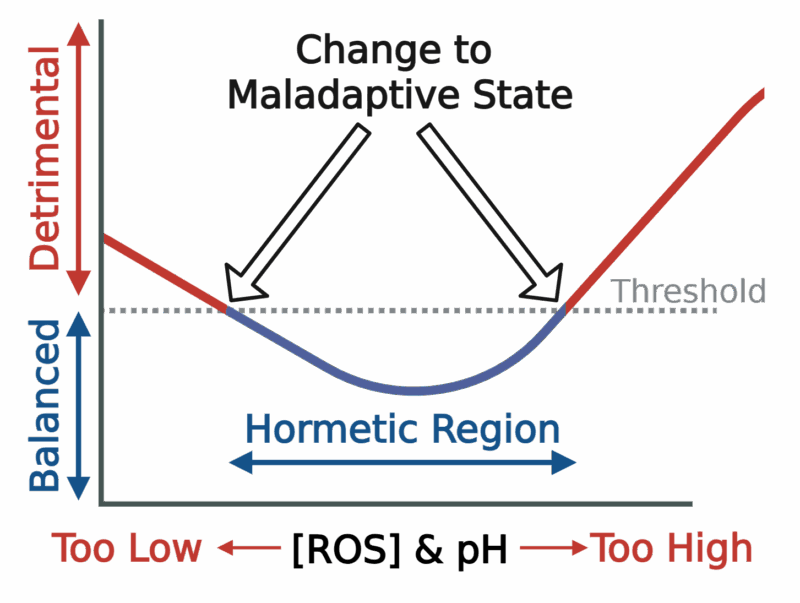
Figure 1: The hormetic window of ROS and pH
From Subtle Imbalance to Chronic Disease
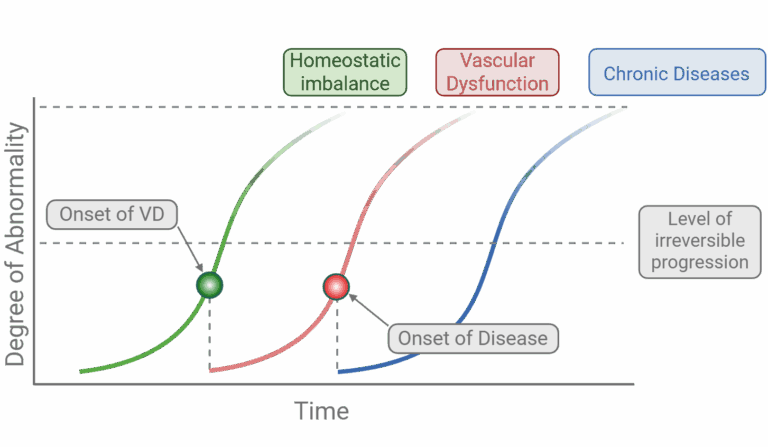
How quickly does a transient imbalance turn into disease? Plotting degree of abnormality against time reveals a predictable trajectory. A protracted, sub-clinical phase of homeostatic imbalance (green) silently deteriorates the endothelium until a tipping point—the onset of vascular dysfunction (VD)—is reached. If left unchecked, the same molecular drivers (ROS and pH dys-regulation) accelerate (red) toward an irreversible threshold that paves the way for chronic neurovascular and neurodegenerative disorders (blue). By intervening early—before the red inflection—disease progression may still be reversible.
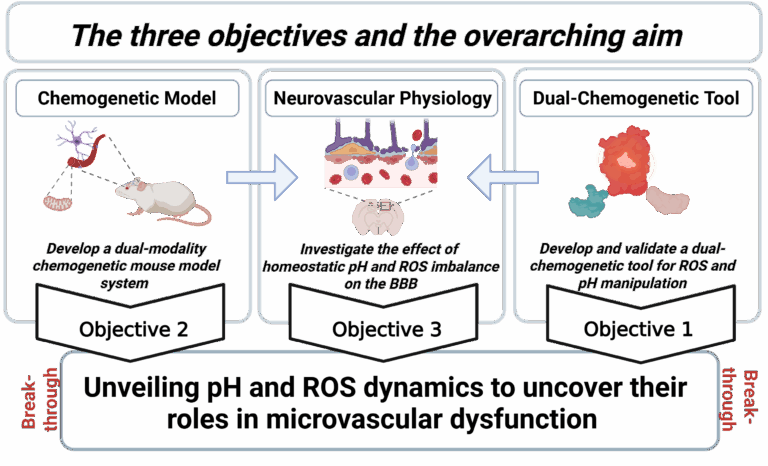
Our Three-Pronged Research Strategy
To convert these concepts into actionable biology we pursue three integrated objectives:
|
Objective |
What we build |
Why it matters |
|
1 – Dual-chemogenetic tool |
A single, genetically encoded construct that allows orthogonal and titratable control of ROS and pH in living cells and tissues |
Provides the precision needed to map causality instead of correlation |
|
2-Chemogenetic mouse model |
A dual-modality transgenic line combined with site-directed substrate delivery (cOFM) |
Enables region-specific perturbation of the neurovascular unit in vivo |
|
3–Neurovascular physiology |
Multimodal imaging, multi-omics and functional assays to quantify barrier integrity, signalling and metabolism under defined ROS/pH states |
Links molecular changes to blood-brain-barrier (BBB) function and behaviour |
Together these aims converge on a single mission:
“Unveiling pH and ROS dynamics to uncover their roles in microvascular dysfunction.”
What this research means for the scientific community
Mechanistic insight – By manipulating two fundamental cues in real time, we can dissect how endothelial cells integrate metabolic, redox and proton signals.
Translational potential – Pinpointing the earliest reversible steps in Figure 2 offers a window for therapeutic intervention before chronic damage ensues.
Enabling technology – The dual-chemogenetic toolkit (Figure 3) will be openly shared, empowering other researchers to interrogate redox and pH biology across organ systems.
